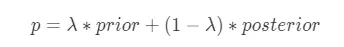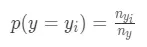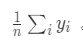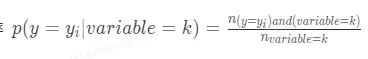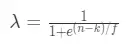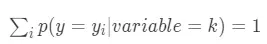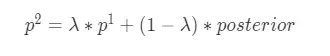一 前言
對於一個類別特徵,如果這個特徵的取值非常多,則稱它為高基數(high-cardinality)類別特徵。在深度學習場景中,對於類別特徵我們一般採用Embedding的方式,通過預訓練或直接訓練的方式將類別特徵值編碼成向量。在經典機器學習場景中,對於有序類別特徵,我們可以使用LabelEncoder進行編碼處理,對於低基數無序類別特徵(在lightgbm中,默認取值個數小於等於4的類別特徵),可以採用OneHotEncoder的方式進行編碼,但是對於高基數無序類別特徵,若直接採用OneHotEncoder的方式編碼,在目前效果比較好的GBDT、Xgboost、lightgbm等樹模型中,會出現特徵稀疏性的問題,造成維度災難, 若先對類別取值進行聚類分組,然後再進行OneHot編碼,雖然可以降低特徵的維度,但是聚類分組過程需要藉助較強的業務經驗知識。本文介紹一種針對高基數無序類別特徵非常有效的預處理方法:平均數編碼(Mean Encoding)。在很多數據挖掘類競賽中,有許多人使用這種方法取得了非常優異的成績。
二 原理
平均數編碼,有些地方也稱之為目標編碼(Target Encoding),是一種基於目標變量統計(Target Statistics)的有監督編碼方式。該方法基於貝葉斯思想,用先驗概率和後驗概率的加權平均值作為類別特徵值的編碼值,適用於分類和迴歸場景。平均數編碼的公式如下所示:
其中:
1. prior為先驗概率,在分類場景中表示樣本屬於某一個\_y\_\_i_的概率
其中\_n\_\_y\_\_i\_表示y =\_y\_\_i_時的樣本數量,\_n\_\_y_表示y的總數量;在迴歸場景下,先驗概率為目標變量均值:
2. posterior為後驗概率,在分類場景中表示類別特徵為k時樣本屬於某一個\_y\_\_i_的概率
在迴歸場景下表示 類別特徵為k時對應目標變量的均值。
3. _λ_為權重函數,本文中的權重函數公式相較於原論文做了變換,是一個單調遞減函數,函數公式:
其中 輸入是特徵類別在訓練集中出現的次數n,權重函數有兩個參數:
① k:最小閾值,當n = k時,_λ_= 0.5,先驗概率和後驗概率的權重相同;當n < k時,_λ_\> 0.5, 先驗概率所佔的權重更大。
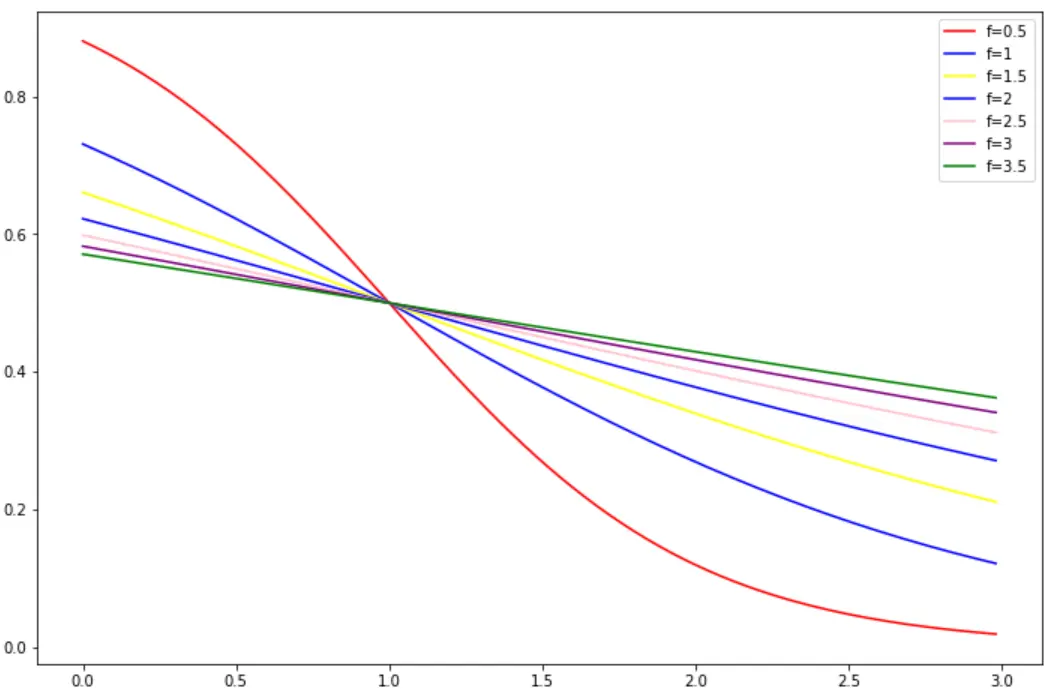
② f:平滑因子,控制權重函數在拐點處的斜率,f越大,曲線坡度越緩。下面是k=1時,不同f對於權重函數的影響:
由圖可知,f越大,權重函數S型曲線越緩,正則效應越強。
對於分類問題,在計算後驗概率時,目標變量有C個類別,就有C個後驗概率,且滿足
一個 \_y\_\_i_ 的概率值必然和其他 \_y\_\_i_ 的概率值線性相關,因此為了避免多重共線性問題,採用平均數編碼後數據集將增加C-1列特徵。對於迴歸問題,採用平均數編碼後數據集將增加1列特徵。
三 實踐
平均數編碼不僅可以對單個類別特徵編碼,也可以對具有層次結構的類別特徵進行編碼。比如地區特徵,國家包含了省,省包含了市,市包含了街區,對於街區特徵,每個街區特徵對應的樣本數量很少,以至於每個街區特徵的編碼值接近於先驗概率。平均數編碼通過加入不同層次的先驗概率信息解決該問題。下面將以分類問題對這兩個場景進行展開:
1. 單個類別特徵編碼:
在具體實踐時可以藉助category_encoders包,代碼如下:
import pandas as pd
from category_encoders import TargetEncoder
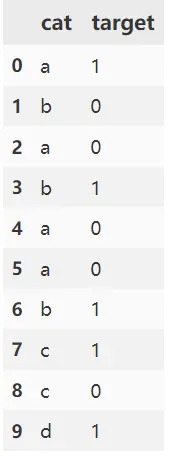
df = pd.DataFrame({'cat': ['a', 'b', 'a', 'b', 'a', 'a', 'b', 'c', 'c', 'd'],
'target': [1, 0, 0, 1, 0, 0, 1, 1, 0, 1]})
te = TargetEncoder(cols=["cat"], min_samples_leaf=2, smoothing=1)
df["cat_encode"] = te.transform(df)["cat"]
print(df)
# 結果如下:
cat target cat_encode
0 a 1 0.279801
1 b 0 0.621843
2 a 0 0.279801
3 b 1 0.621843
4 a 0 0.279801
5 a 0 0.279801
6 b 1 0.621843
7 c 1 0.500000
8 c 0 0.500000
9 d 1 0.634471
2. 層次結構類別特徵編碼:
對以下數據集,方位類別特徵具有{'N': ('N', 'NE'), 'S': ('S', 'SE'), 'W': 'W'}層級關係,以compass中類別NE為例計算\_y\_\_i_=1,k = 2 f = 2時編碼值,計算公式如下:
其中\_p\_1為HIER\_compass\_1中類別N的編碼值,計算可以參考單個類別特徵編碼: 0.74527,posterior=3/3=1,_λ_= 0.37754 ,則類別NE的編碼值:0.37754 0.74527 + (1 - 0.37754) 1 = 0.90383。
代碼如下:
from category_encoders import TargetEncoder
from category_encoders.datasets import load_compass
X, y = load_compass()
# 層次參數hierarchy可以為字典或者dataframe
# 字典形式
hierarchical_map = {'compass': {'N': ('N', 'NE'), 'S': ('S', 'SE'), 'W': 'W'}}
te = TargetEncoder(verbose=2, hierarchy=hierarchical_map, cols=['compass'], smoothing=2, min_samples_leaf=2)
# dataframe形式,HIER_cols的層級順序由頂向下
HIER_cols = ['HIER_compass_1']
te = TargetEncoder(verbose=2, hierarchy=X[HIER_cols], cols=['compass'], smoothing=2, min_samples_leaf=2)
te.fit(X.loc[:,['compass']], y)
X["compass_encode"] = te.transform(X.loc[:,['compass']])
X["label"] = y
print(X)
# 結果如下,compass_encode列為結果列:
index compass HIER_compass_1 compass_encode label
0 1 N N 0.622636 1
1 2 N N 0.622636 0
2 3 NE N 0.903830 1
3 4 NE N 0.903830 1
4 5 NE N 0.903830 1
5 6 SE S 0.176600 0
6 7 SE S 0.176600 0
7 8 S S 0.460520 1
8 9 S S 0.460520 0
9 10 S S 0.460520 1
10 11 S S 0.460520 0
11 12 W W 0.403328 1
12 13 W W 0.403328 0
13 14 W W 0.403328 0
14 15 W W 0.403328 0
15 16 W W 0.403328 1
注意事項:
採用平均數編碼,容易引起過擬合,可以採用以下方法防止過擬合:
- 增大正則項f
- k折交叉驗證
以下為自行實現的基於k折交叉驗證版本的平均數編碼,可以應用於二分類、多分類、迴歸場景中對單一類別特徵或具有層次結構類別特徵進行編碼,該版本中用prior對unknown類別和缺失值編碼。
from itertools import product
from category_encoders import TargetEncoder
from sklearn.model_selection import StratifiedKFold, KFold
class MeanEncoder:
def __init__(self, categorical_features, n_splits=5, target_type='classification',
min_samples_leaf=2, smoothing=1, hierarchy=None, verbose=0, shuffle=False,
random_state=None):
"""
Parameters
----------
categorical_features: list of str
the name of the categorical columns to encode.
n_splits: int
the number of splits used in mean encoding.
target_type: str,
'regression' or 'classification'.
min_samples_leaf: int
For regularization the weighted average between category mean and global mean is taken. The weight is
an S-shaped curve between 0 and 1 with the number of samples for a category on the x-axis.
The curve reaches 0.5 at min_samples_leaf. (parameter k in the original paper)
smoothing: float
smoothing effect to balance categorical average vs prior. Higher value means stronger regularization.
The value must be strictly bigger than 0. Higher values mean a flatter S-curve (see min_samples_leaf).
hierarchy: dict or dataframe
A dictionary or a dataframe to define the hierarchy for mapping.
If a dictionary, this contains a dict of columns to map into hierarchies. Dictionary key(s) should be the column name from X
which requires mapping. For multiple hierarchical maps, this should be a dictionary of dictionaries.
If dataframe: a dataframe defining columns to be used for the hierarchies. Column names must take the form:
HIER_colA_1, ... HIER_colA_N, HIER_colB_1, ... HIER_colB_M, ...
where [colA, colB, ...] are given columns in cols list.
1:N and 1:M define the hierarchy for each column where 1 is the highest hierarchy (top of the tree). A single column or multiple
can be used, as relevant.
verbose: int
integer indicating verbosity of the output. 0 for none.
shuffle : bool, default=False
random_state : int or RandomState instance, default=None
When `shuffle` is True, `random_state` affects the ordering of the
indices, which controls the randomness of each fold for each class.
Otherwise, leave `random_state` as `None`.
Pass an int for reproducible output across multiple function calls.
"""
self.categorical_features = categorical_features
self.n_splits = n_splits
self.learned_stats = {}
self.min_samples_leaf = min_samples_leaf
self.smoothing = smoothing
self.hierarchy = hierarchy
self.verbose = verbose
self.shuffle = shuffle
self.random_state = random_state
if target_type == 'classification':
self.target_type = target_type
self.target_values = []
else:
self.target_type = 'regression'
self.target_values = None
def mean_encode_subroutine(self, X_train, y_train, X_test, variable, target):
X_train = X_train[[variable]].copy()
X_test = X_test[[variable]].copy()
if target is not None:
nf_name = '{}_pred_{}'.format(variable, target)
X_train['pred_temp'] = (y_train == target).astype(int) # classification
else:
nf_name = '{}_pred'.format(variable)
X_train['pred_temp'] = y_train # regression
prior = X_train['pred_temp'].mean()
te = TargetEncoder(verbose=self.verbose, hierarchy=self.hierarchy,
cols=[variable], smoothing=self.smoothing,
min_samples_leaf=self.min_samples_leaf)
te.fit(X_train[[variable]], X_train['pred_temp'])
tmp_l = te.ordinal_encoder.mapping[0]["mapping"].reset_index()
tmp_l.rename(columns={"index":variable, 0:"encode"}, inplace=True)
tmp_l.dropna(inplace=True)
tmp_r = te.mapping[variable].reset_index()
if self.hierarchy is None:
tmp_r.rename(columns={variable: "encode", 0:nf_name}, inplace=True)
else:
tmp_r.rename(columns={"index": "encode", 0:nf_name}, inplace=True)
col_avg_y = pd.merge(tmp_l, tmp_r, how="left",on=["encode"])
col_avg_y.drop(columns=["encode"], inplace=True)
col_avg_y.set_index(variable, inplace=True)
nf_train = X_train.join(col_avg_y, on=variable)[nf_name].values
nf_test = X_test.join(col_avg_y, on=variable).fillna(prior, inplace=False)[nf_name].values
return nf_train, nf_test, prior, col_avg_y
def fit(self, X, y):
"""
:param X: pandas DataFrame, n_samples * n_features
:param y: pandas Series or numpy array, n_samples
:return X_new: the transformed pandas DataFrame containing mean-encoded categorical features
"""
X_new = X.copy()
if self.target_type == 'classification':
skf = StratifiedKFold(self.n_splits, shuffle=self.shuffle, random_state=self.random_state)
else:
skf = KFold(self.n_splits, shuffle=self.shuffle, random_state=self.random_state)
if self.target_type == 'classification':
self.target_values = sorted(set(y))
self.learned_stats = {'{}_pred_{}'.format(variable, target): [] for variable, target in
product(self.categorical_features, self.target_values)}
for variable, target in product(self.categorical_features, self.target_values):
nf_name = '{}_pred_{}'.format(variable, target)
X_new.loc[:, nf_name] = np.nan
for large_ind, small_ind in skf.split(y, y):
nf_large, nf_small, prior, col_avg_y = self.mean_encode_subroutine(
X_new.iloc[large_ind], y.iloc[large_ind], X_new.iloc[small_ind], variable, target)
X_new.iloc[small_ind, -1] = nf_small
self.learned_stats[nf_name].append((prior, col_avg_y))
else:
self.learned_stats = {'{}_pred'.format(variable): [] for variable in self.categorical_features}
for variable in self.categorical_features:
nf_name = '{}_pred'.format(variable)
X_new.loc[:, nf_name] = np.nan
for large_ind, small_ind in skf.split(y, y):
nf_large, nf_small, prior, col_avg_y = self.mean_encode_subroutine(
X_new.iloc[large_ind], y.iloc[large_ind], X_new.iloc[small_ind], variable, None)
X_new.iloc[small_ind, -1] = nf_small
self.learned_stats[nf_name].append((prior, col_avg_y))
return X_new
def transform(self, X):
"""
:param X: pandas DataFrame, n_samples * n_features
:return X_new: the transformed pandas DataFrame containing mean-encoded categorical features
"""
X_new = X.copy()
if self.target_type == 'classification':
for variable, target in product(self.categorical_features, self.target_values):
nf_name = '{}_pred_{}'.format(variable, target)
X_new[nf_name] = 0
for prior, col_avg_y in self.learned_stats[nf_name]:
X_new[nf_name] += X_new[[variable]].join(col_avg_y, on=variable).fillna(prior, inplace=False)[
nf_name]
X_new[nf_name] /= self.n_splits
else:
for variable in self.categorical_features:
nf_name = '{}_pred'.format(variable)
X_new[nf_name] = 0
for prior, col_avg_y in self.learned_stats[nf_name]:
X_new[nf_name] += X_new[[variable]].join(col_avg_y, on=variable).fillna(prior, inplace=False)[
nf_name]
X_new[nf_name] /= self.n_splits
return X_new
四 總結
本文介紹了一種對高基數類別特徵非常有效的編碼方式:平均數編碼。詳細的講述了該種編碼方式的原理,在實際工程應用中有效避免過擬合的方法,並且提供了一個直接上手的代碼版本。
作者:京東保險 趙風龍
來源:京東雲開發者社區 轉載請註明來源